Research
One of the most profound and intriguing questions in biology concerns the relationships between genetic diversity, morphology and function. This complex biology arises during embryonic development. The key to understanding development and differentiation will be found in uncovering molecular mechanisms of gene regulation and deciphering the regulatory circuitry underlying pluripotency, lineage commitment, regional specification and patterning. Understanding these relationships is essential to combat congenital disease to develop the potential of regenerative medicine.
Cardiovascular disease, but also congenital heart disease, is a top-ranking cause of death. In our group we focus on the dynamic changes in heart cells across the different time spans of embryonic development, adult life (disease), and evolution. We examine heart development in different species (vertebrates, cephalopods) and human stem cell-based models to understand fundamental principles of developmental competence and self-organization, and the relationship between evolution and development. Deep conservation and differences among species are equally instructive to understand the heart better. With clinical partners, we also study tissue remodeling in human heart disease. By studying chromatin state, and gene regulation, and single cell trajectories in human and mouse stem cells and Xenopus embryos, we hope aim to uncover fundamental biological principles and discover new avenues for treating diseases.
Epigenetics of cell identity and lineage commitment
What defines cell identity and how can cells change their properties? Important transitions include the exit of pluripotency and lineage commitment, multi-lineage interactions and cellular maturation. We study these mechanisms using single cell transcriptomic profiling, chromatin immunoprecipitation (ChIP-seq), chromatin accessibility assays (ATAC-seq), and mass spectrometry. In combination with gene perturbation experiments we aim to uncover how chromatin state contributes to transitions in cellular identity and how this is regulated at the molecular level. One particular mechanism we are interested in involves the targeting of chromatin-modifying enzymes to specific genes in the genome. For example, the Polycomb PRC2 complex is a complex of proteins involved in repression of cell lineage identity genes. This complex prevents ectopic expression of master regulators outside their lineage.
Gene-regulatory networks
Based on the conceptually simple premise that transcription factors (TFs) bind their cognate motifs in regions with accessible chromatin, it is possible to construct gene-regulatory networks by integrating results from ATAC-seq, RNA-seq and underlying genomic sequence. We are using these networks to model gene regulatory dynamics, spatial gene regulation and developmental transitions. In combination with single cell analyses of cell populations it can be inferred which regulatory factors play important roles in cellular transitions.
Evolution and development
How do regulatory networks compare between species? Much can be learned from the deeply conserved master control switches of development and differentiation. Such comparative approaches in the analysis of regulatory elements can not only provide information on the conservation of gene regulation, but also on the extent to which gene regulation is rewired in different systems or species. We are exploring heart development in squid, which like all coleoid cephalopods (octopus, squid, cuttlefish) has not one but three hearts. We examine how the three hearts of squid develop from embryonic primordia, how the cardiogenic regulatory networks have been rewired, and how the cell types of the heart have evolved.
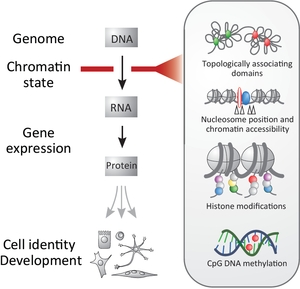
Chromatin state, as defined by chromosome topology, chromatin accessibility and epigenetic modifications, can act as a filter of genomic information influencing lineage commitment and epigenetic stability (Perino and Veenstra, 2016)

TBP family-insensitive network during early development (Gazdag et al., 2016)

Comparison of the gene-regulatory landscape of the two X.laevis hoxb4 homeologs at an early gastrula stage (Session et al., 2016)
