Xenopus tropicalis ChIP-on-chip microarray designs
Whole genome and promoter genome tilepath designs
Chromatin immunoprecipitation (ChIP) combined with genome tile path microarrays (ChIP-chip) can be used to study genome-wide epigenetic profiles and the transcription factor binding repertoire.
Two Roche NimbleGen genome tile path ChIP-chip designs for interrogating the Xenopus tropicalis genome have been developed:
(1) A whole-genome microarray design (5-array set), and
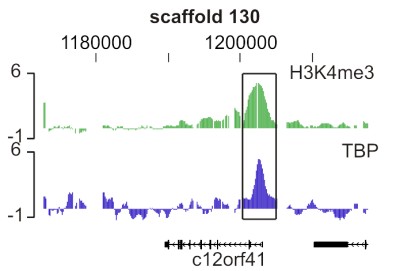
(2) an experimentally defined promoter array, based on 5' ends of transcription units decorated with histone H3 lysine 4 trimethylation in gastrula stage embryos.
These microarray designs can be used to study epigenetic phenomena and transcription factor binding in developing Xenopus embryos.
Publications
Robert C. Akkers*, Simon J. van Heeringen*, J. Robert Manak, Roland D. Green, Hendrik G. Stunnenberg and Gert Jan C. Veenstra. 2010. ChIP-chip designs to interrogate the genome of Xenopus embryos for transcription factor binding and epigenetic regulation. PLoS ONE, 5, e8820 (doi:10.1371/journal.pone.0008820).
 PLoS ONE Open Access
PLoS ONE Open Access
Downloads
The microarray designs can be downloaded as compressed NimbleGen design files (ndf) from the table below. These are optimized designs (v2) which include additional control probes. In addition, the small scaffolds were distributed over the five arrays of the genome-wide 5-array tile path design.
WG: Whole genome 5-array set covering the complete X.tropicalis genome, with the exclusion of of repeat-masked regions. A separate design file for each microarray (1-5).
Promoter: Single array design covering genomic regions enriched for H3K4me3 in gastrula stage embryos (i.e. experimentally verified active promoters), in addition to 5' regions of all annotated genes (JGI FilteredModels, v1).
Genome assembly: Joint Genome Institute version 4.1 (xenTro2, August 2005).
| |||||||||||||||||||||||
|
|
|||||||||||||||||||||||
The microarray data and the earlier version of the microarray designs (v1) have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE19413.
